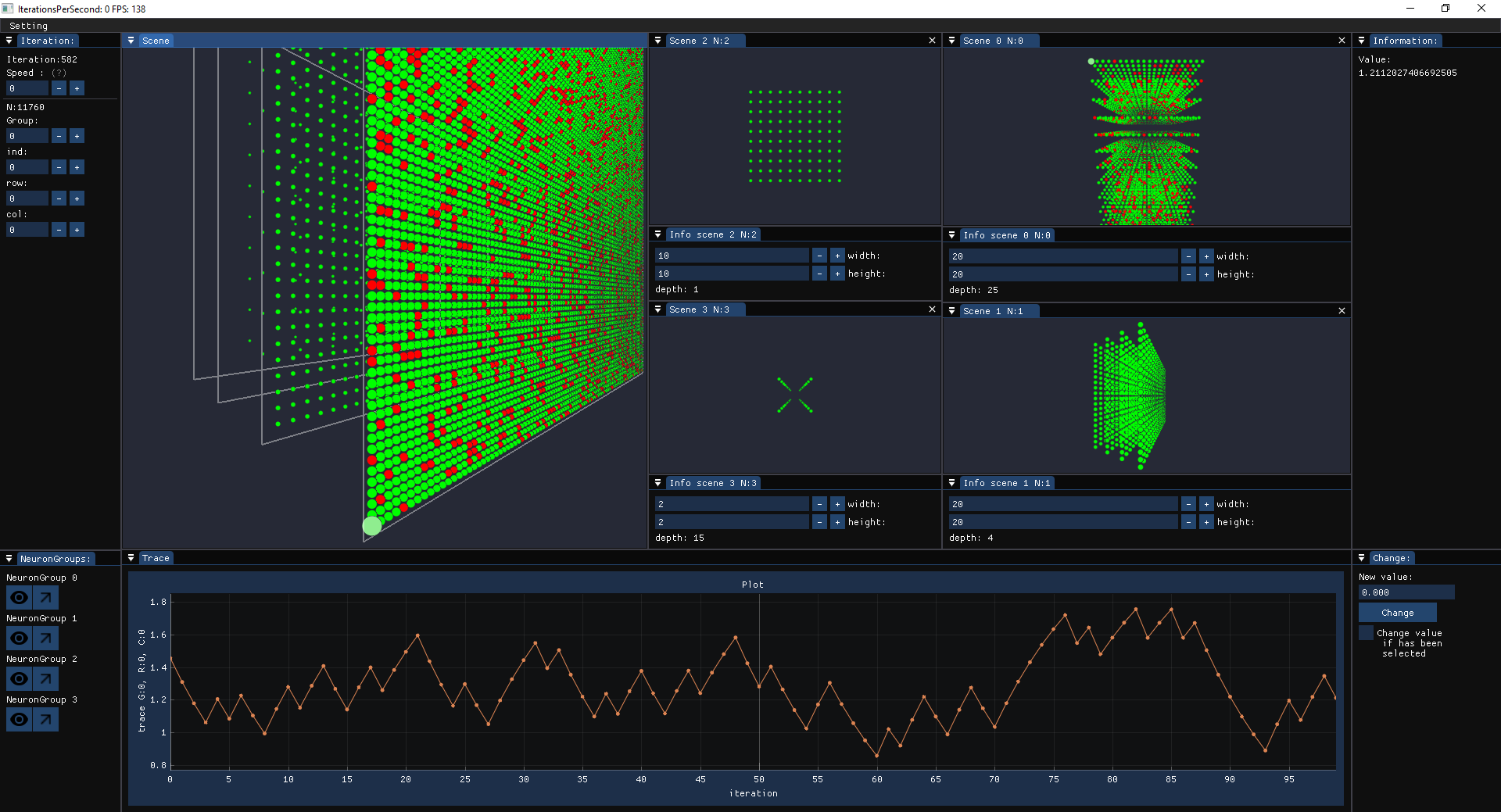
For using visualization first initialize network:
net = Network(dtype=torch.float32,device='cuda')
.
.
.
net.initialize()Then import GUI class:
from visualizer.Visualize_OpenGL_IMGUI import GUIYou can choose to run network and visualizer on a single thread or two seprate threads.
For single thread use this code:
GUI(net).initializeOpenGL()For two thread use this code (recommended):
GUI(net).MultiThreadRun()example_gui_with_raster.py is simple network with GUI with raster plots:
Note:
Networks on both the CPU and GPU (CUDA) can be visualized. Moreover, there is no need to transfer data between the CPU and GPU for visualizing GPU networks, except for their plots (e.g., raster and trace).

PymoNNtorch is a Pytorch-adapted version of PymoNNto.
- Free software: MIT license
- Documentation: https://pymonntorch.readthedocs.io.
- Use
torchtensors and Pytorch-like syntax to create a spiking neural network (SNN). - Simulate an SNN on CPU or GPU.
- Define dynamics of SNN components as
Behaviormodules. - Control over the order of applying different behaviors in each simulation time step.
You can use the same syntax as PymoNNto to create you network:
from pymonntorch import *
net = Network()
ng = NeuronGroup(net=net, tag="my_neuron", size=100, behavior=None)
SynapseGroup(src=ng, dst=ng, net=net, tag="recurrent_synapse")
net.initialize()
net.simulate_iterations(1000)Similarly, you can write your own Behavior Modules with the same logic as PymoNNto; except using torch tensors instead of numpy ndarrays.
from pymonntorch import *
class BasicBehavior(Behavior):
def initialize(self, neurons):
super().initialize(neurons)
neurons.voltage = neurons.vector(mode="zeros")
self.threshold = 1.0
def forward(self, neurons):
firing = neurons.voltage >= self.threshold
neurons.spike = firing.byte()
neurons.voltage[firing] = 0.0 # reset
neurons.voltage *= 0.9 # voltage decay
neurons.voltage += neurons.vector(mode="uniform", density=0.1)
class InputBehavior(Behavior):
def initialize(self, neurons):
super().initialize(neurons)
for synapse in neurons.afferent_synapses['GLUTAMATE']:
synapse.W = synapse.matrix('uniform', density=0.1)
synapse.enabled = synapse.W > 0
def forward(self, neurons):
for synapse in neurons.afferent_synapses['GLUTAMATE']:
neurons.voltage += synapse.W@synapse.src.spike.float() / synapse.src.size * 10
net = Network()
ng = NeuronGroup(net=net,
size=100,
behavior={
1: BasicBehavior(),
2: InputBehavior(),
9: Recorder(['voltage']),
10: EventRecorder(['spike'])
})
SynapseGroup(ng, ng, net, tag='GLUTAMATE')
net.initialize()
net.simulate_iterations(1000)
import matplotlib.pyplot as plt
plt.plot(net['voltage',0][:, :10])
plt.show()
plt.plot(net['spike.t',0], net['spike.i',0], '.k')
plt.show()This package was created with Cookiecutter and the audreyr/cookiecutter-pypackage project template.
It changes the codebase of PymoNNto to use torch rather than numpy and tensorflow numpy.