| title | output | ||||||||
|---|---|---|---|---|---|---|---|---|---|
EHA Modeling and Analytics Best Practices |
|
Edit this document here.
What madness is this?
This document is to provide an outline of the best practices for project organization and programming we aim for our on the EHA Modeling & Analytics team. In each topic below you'll find a general outline, instructions for how to get set up on our various systems, and links to more resources.
In general, we aim to produce sound analyses that are:
- Self-contained
- Well-documented and communicated
- Easily reproducible
- Sharable
- Version controlled
Together, these attributes help assure that our work is correct, can be built off of and extended, meets requirements for sharing and publication, and can be continued through staff turnover.
The tools we use to accomplish this are mostly, but not exclusively, based around the R programming language and git version control. Other teams at EHA use other tools (e.g., the technology team mostly uses Python and Javascript, much of our work with partners is MS Office based).
The guidelines in the document represent an ideal we aim for but to not always attain. Remember:
- Best practices are always evolving.
- Don't let the perfect be the enemy of the good.
- Other teams and external partners have different workflows and we adjust as neccessary to collaborate.
- Our goal is to do good science to advance conservation and human health, not be slick programmers.
You can find some slides from a previous presentation on this topic here.
The philosophy and guidelines in this document owe an enormous amount to the work of the Software and Data Carpentry and rOpenSci organizations, and the work of Hadley Wickham and Jenny Bryan. You'll find many links to their work below.
Can everything be re-done easily if I change one data point in the inputs?
At EHA R is our primary, though not exclusive, tool for analysis and modeling work. R is not just a piece of software for statistics and data manipulation but a computer language, meaning that analsyes are scripted and thus can be automated, run again, built upon and extended.
Learning R is beyond the scope of this document, and you likely already have some experience in it, but some good starting points are:
-
Swirl, a set of interactive lessons run right in R.
-
R for Data Science by Hadley Wickam is a beginner/intermediate text that we highly recommend for getting up to speed with the particular workflows we recommend and the most recent packages that support them.
-
Advanced R (Wickham) is very good for understanding how the language works.
-
Efficient R by Colin Gillespie and Robin Lovelace is helpful for imporving workflows and speeding up code.
-
R Packages (Wickham) is good for package development.
-
Cheatsheets from RStudio are a useful references for a number of things.
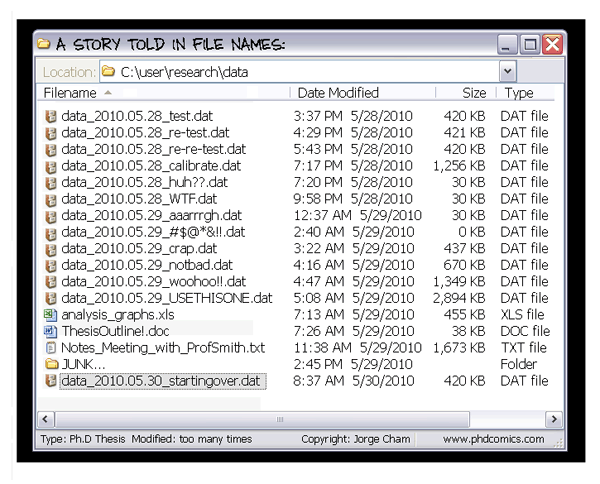
- If I copied this whole folder onto someone else's computer, could they pick up the project?
- Are the folder organization and file naming clear?
We aim to organize projects in a self-contained way, with clear separation between raw data, processed data, exploratory analyses, and final products.
- Some exceptions for large data sets or rapidly changing data sets.
In these cases, data can be organized as a separate folder
or poject.
- In many cases it is actually best for data to be organized as a separate project from analysis. This allows multiple analysis projects to rely on the same upstream data project, avoiding multiple versions of data. In these cases the "data" project should include raw data, aggregation and cleaning, and its output will be cleaned but complete (not summarized data). Analysis projects can import this data as a first step.
- Here is a nice blog post about project structure and a few more alternatives.
- For anything using R, RStudio projects are a good idea for project organization. Here's a Software Carpentry Lesson on RStudio projects.
How do we work together and keep a useful record of our interactions?
-
Slack is our office chat tool and is good for day-to-day communication. Slack does not have to be an instant communication tool - some people prefer to check it a few times a day. Check with your supervisor about your project/team preferences. Slack's main purpose is to organize our communication by channels specific to a topic or project. It is good for keeping information from one project together in a way that can be referenced later by new team members, rather than being lost in various e-mail inboxes. A channel can be linked to many other tools (Dropbox/Google Drive Folder, GitHub Repository), so as to have a central hub for project management. E-mails can be forwarded to a channel.
-
GitHub (see below) has a good issue-tracking system that accompanies each project and can be used for task management and general communication. This ties messages to a specific project and keeps a good long-term record, and can be connected to a slack channel or integrated with e-mail
-
Remember that your Slack and GitHub communications are part of the project and are likely to be seen by internal and external collaborators.
-
ScreenHero is a screen-sharing tool that enables dual control of computer and is part of Slack. It is great for walking through a project with someone remotely or across the office, and also debugging.
- Download and install Slack and ScreenHero. Contact Toph (Tech), or Noam (Modeling & Analytics) to get an account. Join any appropriate channels.
- There's also a mobile Slack app for iOS and Android, which may be helpful if you are traveling.
Will someone understand this thing when I hand it over?
Documentation is essential to collaboration and continuity for our work. Your project should contain documentation to allow a project to be picked up by another user. Documentation includes the following:
- A README document in the top level of the project folder with a high-level explanation of project organization, purpose, and and contact info.
- Metadata for your data set showing its origin and the type of data in each field in a table.
- Comments in your computer code
- Written descriptions of your analyses. The primary medium for this should be R Markdown documents, which allow you to combine code, results, and descriptive text in an easy to update and modify form. Shorter ephermal results can be posted as plots to your project Slack rooms.
- R Markdown is pretty straightforward to learn. You can create your first document and get the basics by going to File > New File > R Markdown in the RStudio menu. When you have time, dive in a bit more with this great lesson on it with accompanying video. Here's an RStudio Reference Sheet for R Markdown.
- (Very optional unless you are asked): ehastyle is our internal EHA R package with R Markdown templates for some reports we produce.
Can the data be shared and published, and easily re-used in other analyses?
- Store data in simple, cross-compatible formats such as CSV files.
- Microsoft Excel can be a useful tool for data entry and organization, but limit its use to that, and organize your data in a way that can be easily exported.
- Metadata! Metadata! Document your data.
- For data sets that cross multiple projects, create data-only project folders for the master version. When these data sets are finalized, they can be deposited in public or private data repositories such as figshare and zenodo. In some cases it makes sense for us to create data-only R packages for easily distributing data internally and externally.
We aim to generally work in a tidy data framework. This approach to structuring data makes iteroperability between tools easier.
- Read Hadley Wickham's tidy data paper for the general concept. Note the packages in this paper are out of date, but the structures and concepts apply.
- R For Data Science is a great online book to read and reference for working in this framework, and gives guidance for the most up-to-date packages (tidyr being the latest analogue of reshape and reshape2).
- Data Carpentry has a Lesson on spreadsheet organization for when you need to do some work in Excel but make it compatible with R.
- Nine simple ways to make it easier to (re)use your data rounds some things out in terms of data sharing. This post is nice, too.
Get the
tidyverse
package for R using install.packages("tidyverse"). This will install
several other relevant packages.
- Can I go back to before I made that mistake?
- Can others see changes others have made to the project and can I see theirs?
Version control is essential to long-term project management and collaboration. We primarily use git for this - we recommend it for any project with more than one file of code. It has a steep learning curve but is very powerful.
-
GitHub is a web service for sharing git-versioned projects that has many great tools for collaboration. We have an organizational GitHub account so we can have private repositories and work in teams with shared projects.
-
For projects with little code-based work, there are other options, as well:
- Google Docs/Word Track Changes are limited to single documents
- Dropbox can track all files in a shared project/folder
- Allows one to view/revert to any previous version of a file in the folder
- Easily sharable
- Does not travel well - history is lost when project moves elsewhere
- File histories are independent - does not track interrelated changes.
-
Avoid filename-based version control:
Git has a steep learning curve and we recommend you spend some time learning rather than only trying to pick it up as you go along.
- Here is a good video course (Part1 / Part2), based on the Software Carpentry curriculum. (Links to the syllabus are in the notes below the videos.)
- Happy Git with R is a great reference for the nuts and bolts of connecting your git, GitHub and R workflows
- Go through the installation steps Happy Git with R's "Installation" and "Connect" chapters and Appendix B
- Note when setting up your GitHub account that one account can have multiple e-mail addresses associated with it, so you can split your work and personal stuff without needing multiple accounts (see here).
- Give Noam (Modeling & Analytics) or Toph (Tech), your GitHub username so they can make you a member of the organizational EHA account and be given access to the appropriate teams.
- Install Dropbox on your computer with your EHA account (note you can have separate personal and EHA Dropbox folders)
- Check that your EHA email gives you access to Google Drive. If you prefer it, or your supervisor specifies it, install it locally on your computer
- Has my work recieved feedback? Has a second set of eyes checked it for correctness?
- Have I learned from my colleagues' work?
Just like any piece of writing that you do, your analysis code should be reviewed by a peer or supervisor. There are generally two types of code reviews we engage in:
- Unit reviews are reviews of discrete, small parts of a project. This might be an analysis that you took a few days or a couple of weeks to complete, and consists of 1-2 files or a few dozen to hundred lines of code. When you complete such a discrete unit, you should solicit feedback.
- Project reviews are reviews of a whole project as it wraps up, such as prior to the submission of a manuscript. These reviews aim to check that the project is complete, understandable and reproducible.
Reviews can be either
- In person reviews where you go over your code with your team or at our informal science meetings. ScreenHero can also be used for this.
- Written reviews where a peers place comments in your code or use the commenting and reviewing features on GitHub.
or both.
- Check out Fernando Perez's tips for code review in the lab.
- Read the Mozilla Guide to Code Review in the Lab
- Check out some rOpenSci package review examples to look at one kind of code review in action.
- Best practices for this are evolving. Check out a recent conversation among scientists on Twitter on the topic
Is this code doing what I think its doing? Is this data correct
Most code should be accompanied by some form of testing. This scales with the size and type of project. Your work should generally accompanied with testing code or outputs that show that your models behave appropriately, are statisically sound, that your code is running as you expect and your data is checked for quality.
- Test driven data analysis is a neat blog on this subject.
- There's a testing chapter in the R Packages book.
- The vingettes and README files of the packages below are useful.
- R packages: assertr or validate for testing that data meets criteria visdat for visually inspecting tabular data. (though there are many ways to plot your data for inspection). testthat for functions and R packages.
How can I make this giant beast of a model run faster?
- Our Aegypti server has 40 cores and 250G of RAM, and can be accessed from anywhere, and has and easy-to-use RStudio interface. It's a good go-to for most biggish analyses you might do.
- The server is generally most useful if you can parallelize your R code across many processors . It's also useful if you have jobs that need a large amount of memory (often big geospatial analyses), or just something that needs to run all weekend while your computer does other things.
- We have an #eha-servers Slack room, for coordinating use of this and other servers check in there if you have questions or before running a big job.
- We also have accounts for Amazon Web Services for appropriate projects.
- Contact Noam for access to this machine and he will create an account and password for you and give you further instructions.
- Log on to the RStudio server interface by pointing your browser at http://aegypti.ecohealthalliance.org:8787/.
For SSH-based access just use
aegypti.ecohealthalliace.orgfrom the terminal. - You will use GitHub to move project work back and forth from your local machine. You will need to setup GitHub access from this machine using SSH keys as described in Happy Git with R Chapter 12. You'll also have to set up your tokens Appendix B again.
- Chapter 7.4 of Efficient R provides a brief introduction to parallelization.
- Not everything can be done from the RStudio server interface on the server. If you are not familiar with the shell interface, brush up via this Software Carpentry lesson. Look at this chapter from an old version of the lesson for instructions on using the Secure Shell (SSH) to login to the server remotely and setting up keys so you don't have to enter your password every time.
- Other helpful things you might look into are tmux for keeping shell processes running when you log off (already installed), and SSH config files for simplifying shell logon.
How do I make sure that all my software and configurations needed for a project are portable?
-
Packrat or checkpoint to fix R package versions.
-
Docker for everything
-
Makefiles can automate a complex, multipart project. Here's a lesson on them from Software Carpentry
-
R packages can be a useful project output. We have some in-house R packages to provide access to internal data and generate reports, and may be developing more for external audiences. Hadley Wickham's R Packages Book provides guidance for these, and we expect our packages to be up to rOpenSci standards.
How do I solve this problem? How do I get my skills up to snuff?
We have an #r-discuss channel on Slack to ask questions and also news about useful resources and packages. (There's also #python-discuss and #stats-chat.) We prefer that you ask question on this channel rather than privately. This way you draw on the group's knowledge, and everyone can learn from the conversation. In general, if you spend 20 minutes banging your head against your screen trying to figure something out, it's time to ask someone.
Some good questions for the Slack room:
-
Which package should I use for [something]?
-
Anyone have a good reference or tutorial for [package, method]?
-
What does this error mean?
-
W.
T.
actual
F.
Our technology team are a tremendous resource for a number of computing topics (especially web technologies and development operations), but remember that they are our collaborators, not IT support. (We do have straight IT support, mostly for office network issues, through Profound Cloud)
Also, outside EHA:
- Stack Overflow is a popular Q&A site for computer programming that a lot of discussions about R.
- The #rstats hashtag on Twitter is a good place for news and short questions, and general ranting.
If there's a course, workshop, or conference you want to attend to improve these skills, speak with your supervisor, we can often support this.